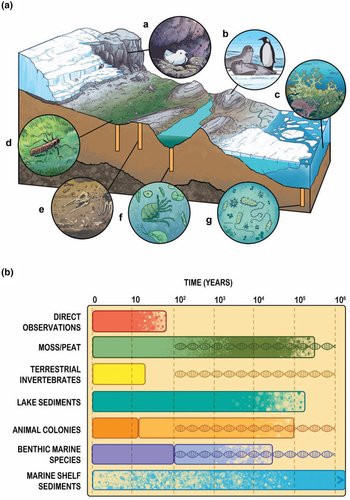
Jan led a paper with co-authors from Securing Antarctica’s Environmental Future (SAEF) to highlight the utility and power of biological archives to understand past ecological and climatic change in Antarctica. Biological archives included extant moss beds and peat profiles, biological proxies in lake and marine sediments, vertebrate animal colonies, and extant terrestrial and benthic marine invertebrates. The paper highlights how emerging biological archives complement other Antarctic paleoclimate archives (e.g. ice cores) by recording the nature and rate of past ecological change, the paleoenvironmental drivers of that change, and constrain current ecosystem and climate models. Significant advances in analytical techniques (e.g., genomics, biogeochemical analyses) have led to new applications and greater power in elucidating the environmental records contained within biological archives. The paper highlights how these emerging biological archives will significantly expand our understanding of past, present, and future ecological change, alongside climate change in Antarctica and at the Southern Ocean.
A video about the article made by GCB: https://twitter.com/GlobalChangeBio/status/1561850857334923269
Link to SAEF article about the paper: https://arcsaef.com/story/accessing-earths-memories/
Link to paper: https://onlinelibrary.wiley.com/doi/10.1111/gcb.16356