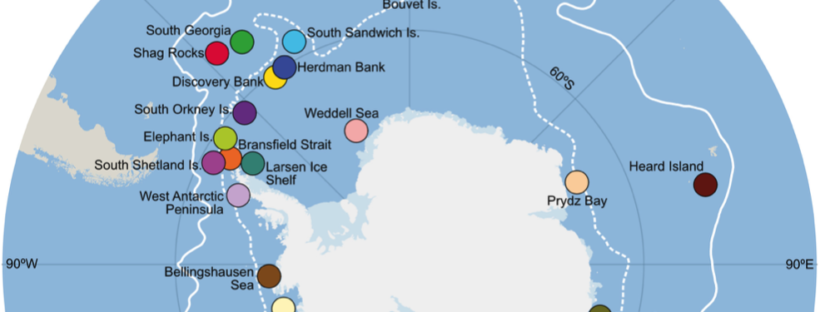
New paper by Sally, Catarina and Jan in Ecology & Evolution, uses COI sequences to investigate the population genetic patterns of the Antarctic brittle stars Ophionotus victoriae and O. hexactis with contrasting life histories (broadcasting vs brooding) and morphology (5 vs 6 arms). They found that, throughout the Pleistocene glacial maxima, O. victoriae likely persisted in deep sea refugia; whereas O. hexactis likely persisted in Antarctic island refugia. This work proposes the evolutionary innovations in O. hexactis (increase in arm number and a switch from broadcast spawning to brooding) could be linked to survival within island refugia, which open up new avenues for future genomic research!
Category: Publications
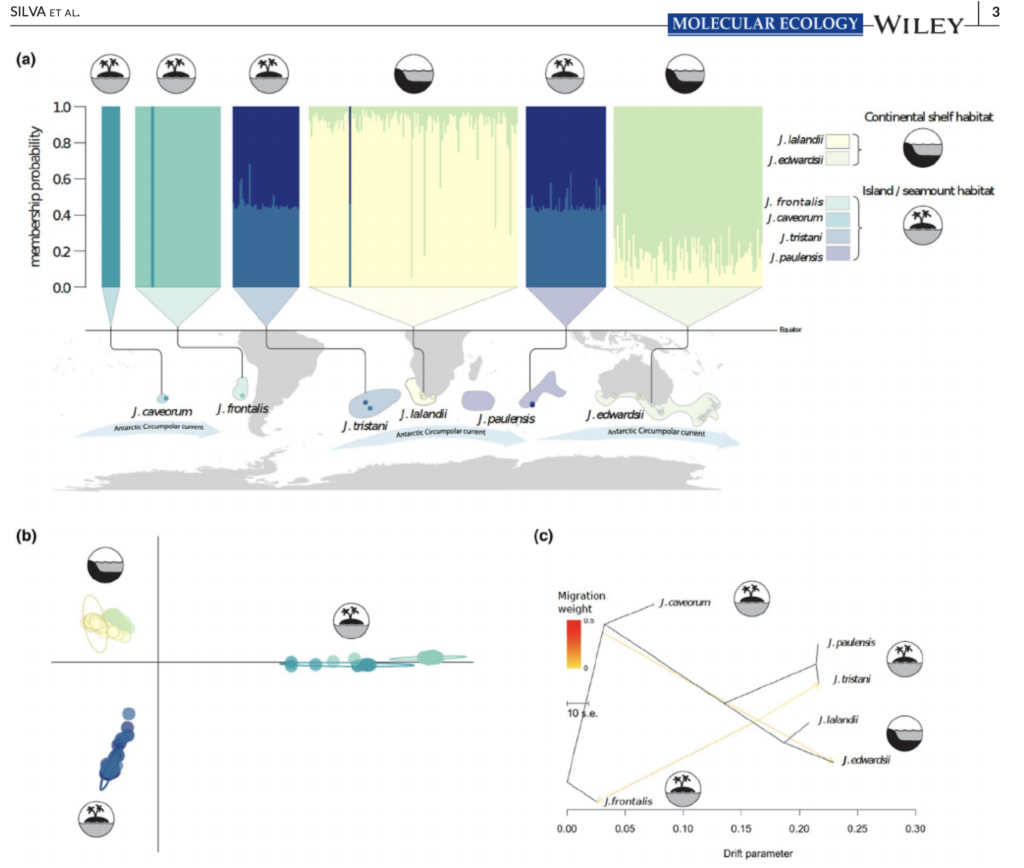
Global drivers of recent diversification in a marine species complex
New paper by Catarina and Jan in Molecular Ecology, (full article here), investigates genome-wide divergence, introgression patterns and inferred demographic history between species pairs of all six extant rock lobster species within the genus Jasus – species with a larval duration of up to two years. Funded by the Australian Research Council, this work shows the important effect of habitat and demographic processes on the recent divergence of species in the genus.
Reference Genome and Population Genomics of coral (Acropora tenuis) on the inshore Great Barrier Reef
New paper by Ira, Jan and Jia in Science Advances (full article here), uses shallow whole genome sequencing to look at demographic history and selection for Acropora tenuis. The main findings are outlined in this tweet thread. Maria Nayfa also made this nice video of Ira explaining why genomes are so useful for understanding the history of the GBR.
ampir a new R package from the group
Antimicrobial peptides are part of the innate immune system and help defend the host against pathogens and regulate the microbiome. Antimicrobial peptides occur in all life, are incredibly diverse, mostly quite small (< 200 amino acids), and only comprise of a small proportion in a genome (~ 1%). This makes them very difficult to find. We created a classification model implemented in an R package, ampir, to predict antimicrobial peptides from protein sequences on a genome-wide scale. ampir was tested on multiple test sets (including complete proteomes) and performed with high accuracy. ampir can be used to narrow down the search space for novel antimicrobial peptides in genomes.
ampir was recently published in Bioinformatics and is available on CRAN and github . Legana has also created a companion repository to accompany the paper and document the thinking behind ampir’s model building process.
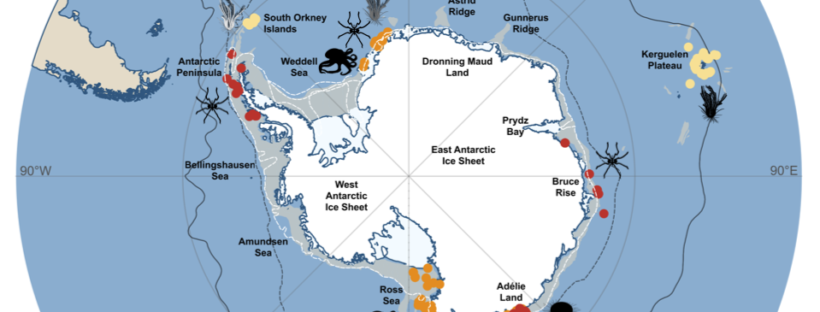
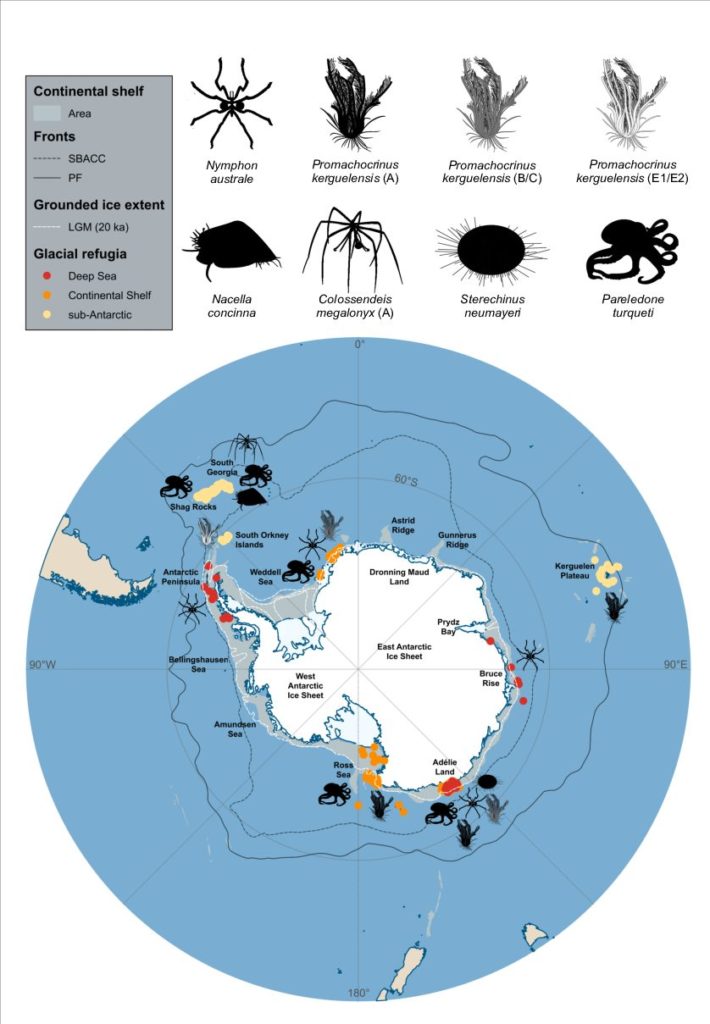
Detecting Glacial Refugia in the Southern Ocean
Throughout the Plio-Pleistocene glacial cycles, the Antarctic ice sheets expanded and contracted repeatedly, which led to the repeated erosion of the Antarctic continental shelf throughout glacial maxima. Even though molecular and paleontological evidence suggests most extant Antarctic benthos persisted in situ in the Southern Ocean during Plio-Pleistocene, where and how they survived remain mostly hypothesised.
Our paper recently published in Ecography synthesises current geological and ecological evidence to understand where and how benthos might have survived glacial cycles and how this challenging period might have impacted past species demography. We also examined current molecular evidence of glacial refugia in the Southern Ocean, as well as discussed future directions for employing testable frameworks and genomic methods in Southern Ocean molecular studies.
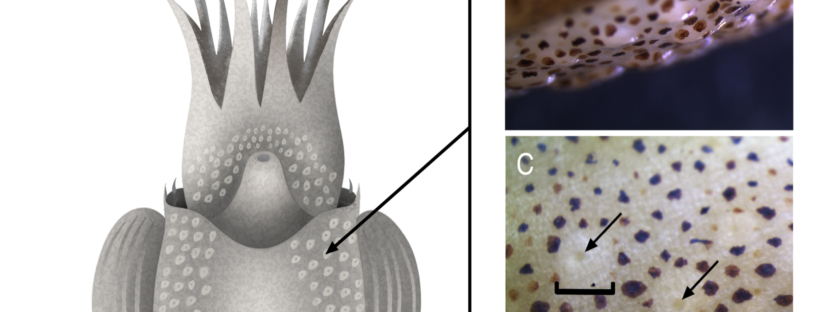
Quantitative Proteomic Analysis of the Slime and Ventral Mantle Glands of the Striped Pyjama Squid (Sepioloidea lineolata)
New paper in Journal of Proteome Research by group alumnus, Nikeisha Caruana uses quantitative proteomics to understand slime secretions in striped pyjama squid. For this work Nikeisha used a multi-tissue comparison to hone in proteins unique to slime-producing glands. The work is also the first time these glands have been described in detail and their proteomic similarity to the slime and physical structure implicates them as likely secretion structures.
Publication Link: https://pubs.acs.org/doi/abs/10.1021/acs.jproteome.9b00738
Nikeisha Caruana is now a research fellow at the Bio 21 institute in Melbourne
Distribution of Palinuridae and Scyllaridae phyllosoma larvae within the East Australian Current: a climate change hot spot
Laura Woodings, Nick Murphy, Andrew Jeffs, Iain Suthers, Geoff Liggins and Jan M. Strugnell
Laura sequenced the barcoding gene, Cytochrome oxidase I (COI), from palinurid and scyllarid lobster larvae (phyllosoma) caught off the east coast of Australia. She detected two tropical and one subtropical palinurid species ~75–1800 km to the south or southwest of their known species distribution. These results indicate tropical lobster species are reaching temperate regions, providing these species the opportunity to establish in temperate regions if or when environmental conditions become amenable to settlement.
Published in Marine and Freshwater Research
Featured image is a Phyllosoma by Ernst Haekel
The evolution and origin of tetrodotoxin acquisition in the blue-ringed octopus (genus Hapalochlaena)
Brooke L. Whitelaw, Ira R. Cooke, Julian Finn, Kyall Zenger, J.M. Strugnell
Blue ringed octopus make use of a deadly neurotoxin, TTX (tetrodotoxin) which is also found a wide variety of other animals. In order to do this they must be able to tolerate the toxin themselves, as well as acquire it from the environment or produce it. This paper reviews what we know about the evolution of ttx acquisition in blue ringed octopuses.
Featured image is by Flickr user Rickard Ling (CC 2.0)
Comparative Proteomic Analysis of Slime from the Striped Pyjama Squid, Sepioloidea lineolata, and the Southern Bottletail Squid, Sepiadarium austrinum (Cephalopoda: Sepiadariidae)
Nikeisha J. Caruana , Jan M. Strugnell, Pierre Faou, Julian Finn, and Ira R. Cooke
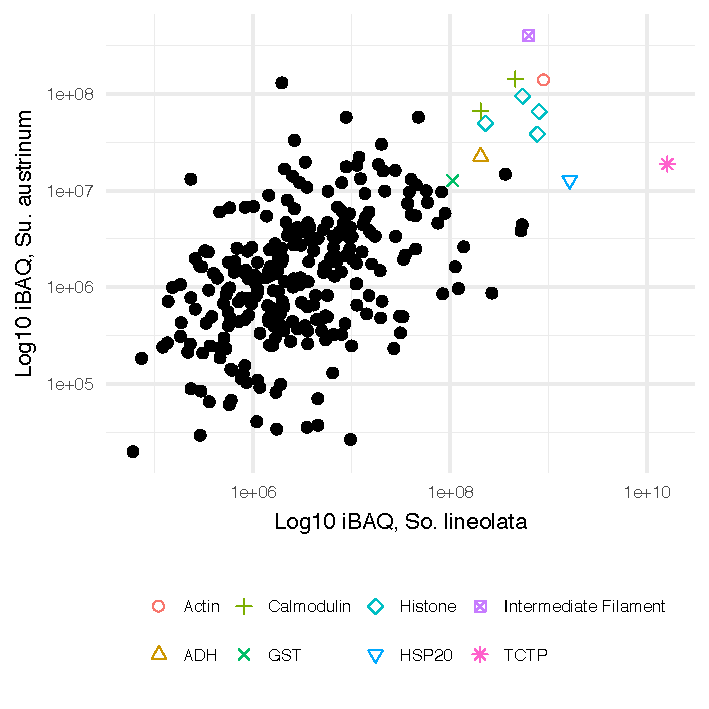
Nikeisha compared the slime from two bobtail squids and found that their proteomic composition was very similar. She also found several highly abundant, short secreted proteins in the slime, for which a function is unknown.
Published in the Journal of Proteome Research
Featured image is of Sepioloidea lineolata. Photograph by Richard Ling <wikipedia@rling.com> [CC BY-SA 3.0], via Wikimedia Commons
Shotgun Proteomics Analysis of Saliva and Salivary Gland Tissue from the Common Octopus Octopus vulgaris
Legana C. H. W. Fingerhut, Jan M. Strugnell, Pierre Faou, Álvaro Roura Labiaga, Jia Zhang, and Ira R. Cooke
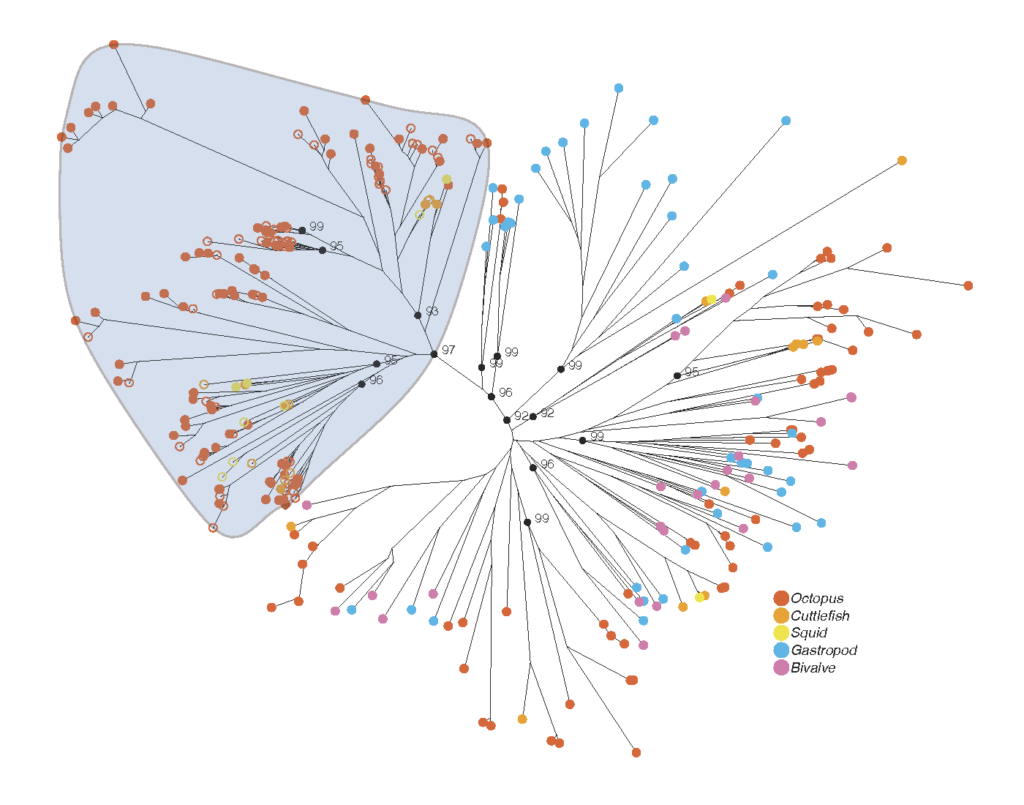
A wide variety of octopus and cuttlefish species use venom to disable and kill their prey. Although this has been known for over a hundred years the molecules responsible for this toxicity remain relatively unknown. In this paper we found that a particular family of proteins (serine proteases) are extremely diverse and abundant in the venom glands of octopus. Although serine proteases are common in all animals (and not always toxins) we found that they are extraordinarily diverse in octopus and cuttlefish, and that this diversity is almost entirely due to molecules that are found in the venom glands.
Most of the work for this paper was done by Legana as part of her honours year. Well done Legana on your first paper.
Featured image is of Felicinda the octopus who was the mother of octopuses featured in this study. Photograph by Álvaro Roura
Published in the Journal of Proteome Research